Cython is an extension of Python, that adds static typing to variables, functions and classes. It combines simplicity of Python and efficiency of C. You can rewrite your code in Cython and compile them to C to achieve higher execution speed.
In this tutorial, you’ll learn how to:
- Install Cython and Compile Cython code
- something about speed
- Write Cython application with statically typed variables and C functions
What Cython is and what it’s used for
The Cython project consists of two parts - a programming language and a compiler. Cython language is a superset of Python that adds support for C types and functions. Cython compiler can understand both languages, but most importantly it can generate very efficient C code from Cython.
Cython was created in 2007 by developers of the Sage computer algebra package, and now it’s popular among scientific users of Python. Its current version is now 0.29 with new version 3 in pre-release. The new version uses Python 3 by default and brings some backward incompatible changes, just like Python 3 brought. In this article you’ll use current stable release 0.29 with Python 3, which it also supports.
One of the main usages of Cython is increasing speed of Python code execution. You rewrite slow parts of your Python code in Cython, compile to fast C code and use it back in Python as an external module.
Installation
Cython needs C completer to be present in the system. It’s installation differs between different operating systems:
Linux
GNU C compiler (gcc) is usually directly available in some distributions or can be easily obtained through a packet manager. On Debian-like systems, like Ubuntu, you can install it with sudo apt-get install build-essential.
Check the installation with gcc -v command.
macOS
C compiler is available as a part of Xcode Command Line Tools. The easiest way to install them is entering xcode-select --install in the Terminal. They are also available to download from https://developer.apple.com/
You can also check the the installation with clang -v command in the Terminal.
Windows
First thing you need to do is to update setuptools - its version must be at least 34.4.0:
pip install --upgrade setuptools
C compilers on Windows are a part of Video Studio framework. You don’t need the whole thing - just install Microsoft Build Tools for Visual Studio 2019.
In the installer select C++ build tools and ensure the latest versions of MSVCv142 - VS 2019 C++ x64/x86 build tools and Windows 10 SDK are checked, like on the screen shot below.
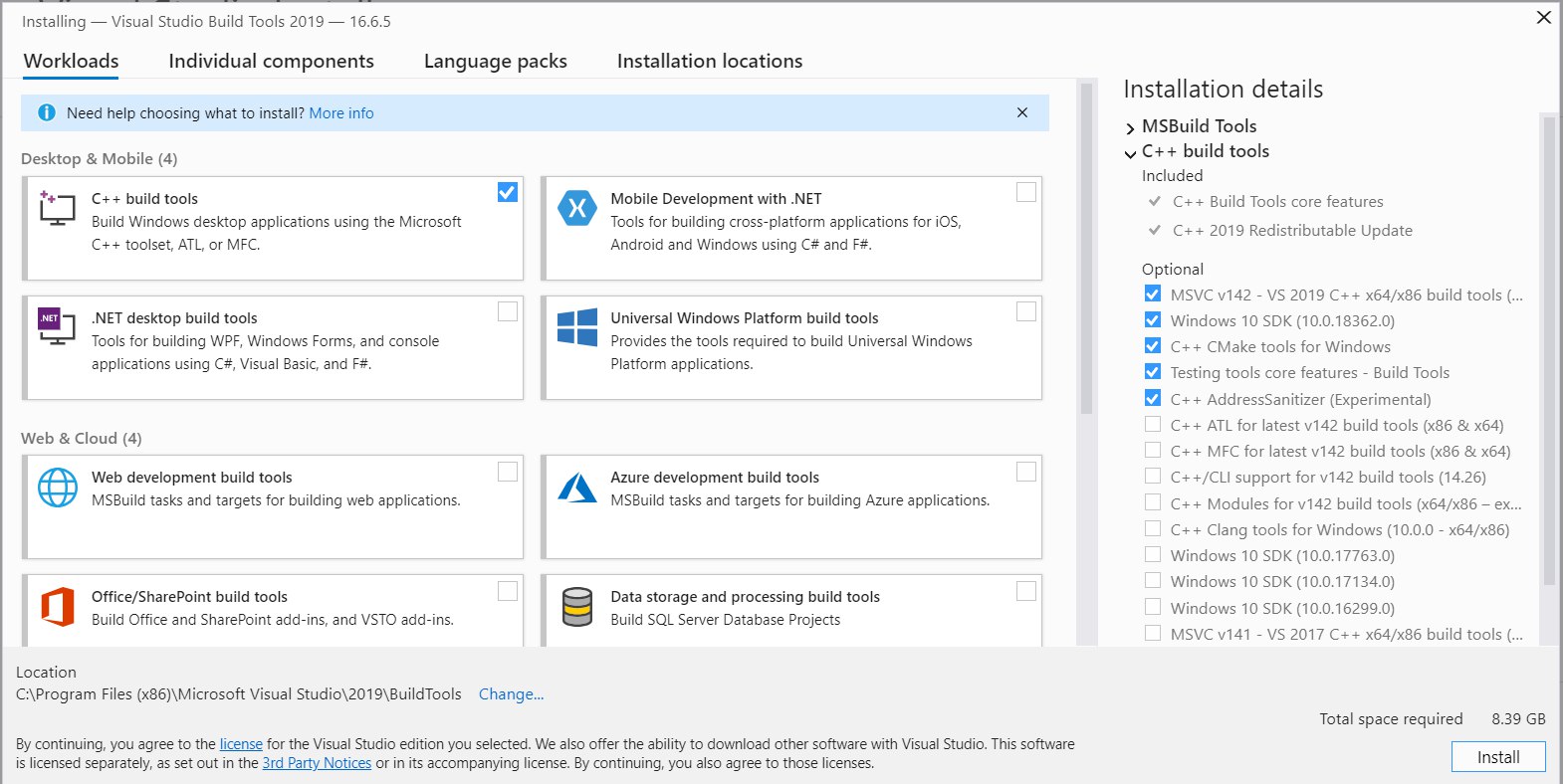
After installation, hit Start and search for Developer Command Prompt. Once it’s open, you can enter cl /? to check whether the installation was successful.
Final step
The simplest way of installing Cython is by using pip:
pip install Cython
If you install in for on continuos integration server, for testing or on platforms for this wheel packages are not provided you may consider an uncompiled version:
pip install Cython --install-option="--no-cython-compile"
Alternatively, you can install Anaconda Python distribution that comes with all the batteries - compiler and Cython installed.
Compilation
Following the tradition, your first application will be a program that prints “Hello, World!”. Cython source files have extension .pyx. Create a new file hello.pyx containing the following code:
def hello():
print("Hello, World!")
The next step is to convert it to C. cython command will read hello.pyx and produce hello.c file:
$ cython -3 hello.pyx
-3 option tells cython to Python 3.
To compile hello.c you’ll need C compiler that is already installed. You need to provide a number of Python- and OS-specific compilation options. On latest Ubuntu 20.04 the command looks like this:
gcc -shared -pthread -fPIC -fwrapv -Wall -O2 -I/usr/include/python3.8/ -o hello.so hello.c
sharedalong withfPICoption instructs the compiler to produced share library, that can by used be another applicationspthreadadds support for multithreadingO2andfwrapvandfno-strict-aliasingturn on C code optimizationsWallenables all compiler warningsIspecifies a path toPython.hheader fileOspecifies output filename- the last positional argument is name of a C source file generated by
cythoncommand
Now you are ready to use newly created shared library in Python. Spin up a Python REPL with python command and type in this code:
>>> import hello
>>> hello.hello()
Hello, World!
Using distutils
There is a more straightforward way to compile Cython without the need to invoke the compiler directly and providing all necessary compilation option. By using standard Python packaging tool distutil and cythonize function from Cython module you can compile hello.pyx directly to shared library.
First, you need to create a setup script. By convention this script is named setup.py. In the first two lines of the script you need to import setup function from distutils and cytonize from Cython:
from distutils.core import setup
from Cython.Build import cythonize
Setup function accepts a number of keyword arguments. First one is a name of our application and the second are extensions to build it with. cytonize accepts a single or multiple - in a form a list - names of Cython source files. The rest of the setup script looks like this:
setup(
name="Hello",
ext_modules = cythonize("hello.pyx", language_level=3),
)
language_level keyword arguments instructs to run cython command with -3 options, enabling Python 3.
In order to compile the application you need to run it with the following command:
python setup.py build_ext --inplace
build_exttellsdisutilsto use extensionsinplaceoption will makehello.sofile to appear in the same directory
Compiling Cython in Jupyter notebook
You can seamlessly use Cython in Jupyter notebooks without any explicit compilation. It may be useful when experimenting with Cython or profiling using many Jupyter’s helpers.
First step is to load a Cython extension:
In [1]: %load_ext Cython
Then in a cell with Cython code start with a magic %%cython like so:
In [2]: %%cython
def hello():
print("Hello, World!")
Now you can call Cython function as you’d call a normal one:
In [3]: hello()
"Hello, World!"
Profiling
Any performance tuning starts with establishing a baseline and finding bottlenecks. Python provides useful utilities to do that including timeit module and cProfile.
Next application you are going to write will calculate sum of squares of all numbers from1 to 1 million.
Create a file sum_of_squares.py and enter the following code in it:
def square(a):
return a**2
def sum_of_squares():
s = 0
for i in range(1, 10**6 + 1):
s += square(i)
return s
if __name__ == "__main__":
print(sum_of_squares())
Python standard library has a nice module called timeit to measure the execution time. You can invoke it from terminal for you application’s source file sum_of_squares.py:
python -m timeit "from sum_of_squares import sum_of_squares; sum_of_squares()"
10 loops, best of 3: 745 msec per loop
This number - 745 msec - will be your baseline.
In order to find where this code spends most time you can use cProfile:
python -m cProfile naive_sum.py
ncalls tottime percall cumtime percall filename:lineno(function)
1 0.000 0.000 0.901 0.901 naive_sum.py:1(<module>)
999999 0.563 0.000 0.563 0.000 naive_sum.py:1(square)
1 0.338 0.338 0.901 0.901 naive_sum.py:4(sum_of_squares)
1 0.000 0.000 0.901 0.901 {built-in method builtins.exec}
1 0.000 0.000 0.000 0.000 {built-in method builtins.print}
1 0.000 0.000 0.000 0.000 {method 'disable' of '_lsprof.Profiler' objects}
Looks like a call to seemingly redundant square function is pretty costly. But don’t jump to refactoring this code right away - let’s see how Cython can help with that.
Firstly, just by compiling it with Cython you can achieve almost 30% increase in performance:
- Rename
sum_of_squares.pytosum_of_squares_cython.pyx - Compile with one of the methods from above
- Use
timeitagain:python -m timeit "from sum_of_squares_cython import sum_of_squares; sum_of_squares()" 10 loops, best of 3: 543 msec per loop
Cython provides an annotation tools that helps with profiling. Each line in the annotation is color coded - darker lines indicate that there is much more C code was generated for them and that they are potentially slower.
To get the annotation you need to call cython command with -a argument on .pyx file:
cython -3 -a naive_sum_cython.pyx
It will produce a sum_of_squares_cython.html file:
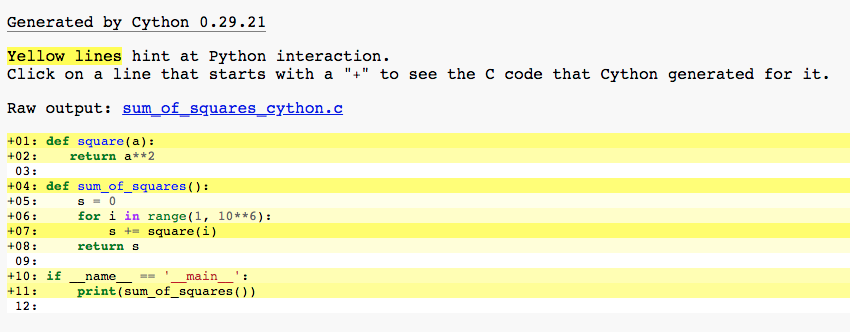
Yellow lines hint at Python interaction.
Click on a line that starts with a “+” to see the C code that Cython generated for it. And again a call to square on line 7 is marked as heavy.
As you can see the program spends a lot of time in a long loop calling a function that does numeric computations. Such “hot” loops with numeric computations are good examples of an code that will benefit from the main feature of Cython - static types.
Defining static types
In Python variables are labels that refer to an object in memory. In any place of your program you can create new variable s and assign an integer to it:
s = 0
The integer 0 is now bounded to the variable s. Later on in the program you can redefine s - assign another value of a different type to s:
s = 'sum of squares'
Python has no problems with dynamically changing the type of a variable - now s points to a string 'sum of squares'.
Variables with static types are more like data containers - they store the value in the variable and their type cannot change.
In Cython to define a variable you need to put cdef keyword and a type before variable name:
cdef int s
You can also initialize the value in the same line:
cdef int s = 0
Let’s see how Cython would react if you try to assign a float to s:
cdef int s = 0
s = 0.0
When compiling the code you’ll get an error:
Cannot assign type 'double' to 'int'
Cython provides all the standard C types, such as char, short, int, long along with Python types of list, dict, tuple, etc. Let’s see how defining a type can speed things up.
A simple loop in Python that sums up numbers may look like this:
def loop():
s = 0
for i in range(1, 10**6+1):
s += i
return s
Timing this function gives: 10 loops, best of 3: 128 msec per loop. But if you define a type for loop index and result variable s:
def loop_with_types():
cdef long s = 0
cdef int i = 0
for i in range(1, 10**6+1):
s += i
return s
the execution time will be 1000 faster: 10 loops, best of 3: 139 ns per loop.
This increase in speed is possible because the loop was converted to pure C and then machine code, while Python code was relying on slow interpreter.
But what about slow square function from sum_of_squares? You can define types for function return and input variables to make them more efficient.
Speeding up functions
You can specify types of the arguments of a Python function by putting a type in front of the argument name:
def square(int a):
return a**2
This function is still a Python function, but with additional type checking. You can also achieve this with using function annotations:
def square(a: int) -> int:
return a**2
cythonize function used in setup.py accepts a parameter annotation_typing that tells Cython to infer type of variables from annotations.
To take advantage from Cython optimizations you need to define a function using cdef and a return type. This, along with defining types for other variables, will give a substantial increase in speed:
cdef long square(int a):
return a**2
def sum_of_squares():
cdef long s = 0
cdef int i = 0
for i in range(1, 10**6 + 1):
s += square(i)
return s
Run time measured with timeit - 10 loops, best of 3: 106 msec per loop - is 5 times faster than before.
But functions defined with cdef are by default available only inside Cython and cannot be imported back to Python. Cython allows to define a functions that is both callable from Python and converted to C functions. If you define a function with cpdef Cython will create 2 versions - Python importable version and a C variant:
cpdef long sum_of_squares():
cdef long s = 0
cdef int i = 0
for i in range(1, 10**6 + 1):
s += square(i)
return s
With having sum_of_squares function like that you lower the run time to 120ns, which is 4 orders of magnitude faster than your baseline of 745msec.
Let’s run Cythin annotation once more on final version of sum_of_squares.pyx:
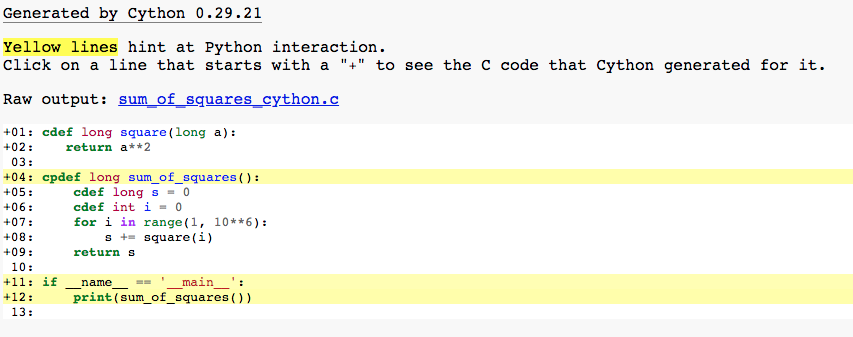 Most of the lines are white, which means that code was optimized.
Most of the lines are white, which means that code was optimized.
Working with arrays
Often numerical computations use arrays. Cython gives access to fast C and NumPy arrays. But there are some important differences when you compare them with standard Python lists.
In Python lists can contain elements of different types. This is a valid list in Python: a = [1, "two", 3.0]. Moreover, you can append items to it anytime.
In Cython, when defining an array, you need to specify a type of elements inside and the length:
cdef double a[10]
This will allocate 80 bytes of memory - every double occupies 8 bytes - and enforce that every element in a is of type double. For more flexibility you can use NumPy arrays.
NumPy arrays are already pretty fast when you use them in regular Python. But Cython can optimize some indexing operations that will make certain operations even faster.
You first need to import NumPy inside Cython. It’s done with a special cimport statement:
cimport numpy as cnp
Now you need to define an arrays with a special syntax:
cdef cnp.ndarray[double, ndim=1] a
The type of variable a is ndarray - same one you use normally with NumPy.
In the square brackets you list a type of arrays elements and a number of dimensions - in this case it’s a one dimensional array.
Let’s find out how using NumPy this way is faster than in Python. It’s a simple function that creates an array with 1000 random elements. Then it adds 1 to each element. The implementation in Python may look like this:
import numpy as np
def numpy_py():
a = np.random.rand(1000)
for i in range(1000):
a[i] += 1
In Cython you define an array first before using it, but the rest of the program is the same:
import numpy as np
cimport numpy as сnp
def numpy_cy():
cdef сnp.ndarray[double, ndim=1] c_arr
a = np.random.rand(1000)
cdef int i
for i in range(1000):
a[i] += 1
Cython version finishes in 21.7 µs vs 954 µs for Python, due to fast access to array element by index operations inside the loop.
Conclusion
You are now able to install and compile Cython code and use it in Python applications. You’ve also learned about profiling tools and now to use them.
You’ve learned:
- how to install Cython on macOS, Linux and Windows
- compile Cython code using 3 different methods
- write simple Cython functions and use statically typed variables
- use fast arrays
